NAMHC Workgroup on High Dimensional Data
Report of Recommendations
June 7, 2024
Table of Contents
I. Roster
Co-Chairs
Laura Almasy, Ph.D.
Professor, Genetics at the Perelman School of Medicine
Department of Biomedical and Health Informatics
University of Pennsylvania
Philadelphia, PA
Damien Fair, PA-C, Ph.D.
Professor
Redleaf Endowed Director of the Masonic Institute of Child Development
Department of Pediatrics
University of Minnesota
Minneapolis, MN
Members
Edwin (Ted) Abel, Ph.D.
Chair and Departmental Executive Officer
Department of Neuroscience and Pharmacology
Director, Iowa Neuroscience Institute
University of Iowa
Iowa City, IA
Laura Jean Bierut, M.D.
Alumni Endowed Professor
Department of Psychiatry
Washington University School of Medicine
St. Louis, MO
Kristen Brennand, Ph.D.
Elizabeth Mears and House Jameson Professor of Psychiatry
Professor of Genetics Department of Psychiatry
Yale University School of Medicine
New Haven, CT
Luca Foschini, Ph.D.
Sage Bionetworks
Seattle, WA
Neda Jahanshad, Ph.D.
Associate Professor of Neurology
Keck School of Medicine
University of Southern California
Los Angeles, CA
Erich D. Jarvis, Ph.D.
Professor, Head of Laboratory
Department of Neurogenetics of Language
Rockefeller University
New York, NY
Robert E. Kass, Ph.D.
Maurice Falk University Professor of Statistics and Computational Neuroscience
Department of Statistics and Data Science, Machine Learning Department and the Neuroscience Institute
Carnegie Mellon University
Pittsburgh, PA
Tuuli Lappalainen, Ph.D.
Professor, Director of the National Genomics Infrastructure and the Genomics Platform of SciLifeLab
KTH-Royal Institute of Technology
Stockholm, Sweden
Senior Associate Faculty Member
New York Genome Center
New York, NY
Cathryn M. Lewis, Ph.D.
Professor of Genetic Epidemiology & Statistics
Head of Department, Social, Genetic Developmental Psychiatry Centre
King’s College London
London, UK
Shannon K. McWeeney, Ph.D.
Professor of Biostatistics and Bioinformatics
Vice Chair, Research
Division of Bioinformatics and Computational Biology, Department of Medical Informatics and Clinical Epidemiology, School of Medicine
Oregon Health & Science University
Portland, OR
Lisa D. Nickerson, Ph.D.
Associate Professor
Harvard Medical School
Director, Applied Neuroimaging Statistics Research Lab
McLean Hospital
Belmont, MA
Laura Scott, M.P.H., Ph.D.
Research Professor
Department of Biostatistics
Center for Statistical Genetics
University of Michigan
Ann Arbor, MI
Members
Erich D. Jarvis, Ph.D.
Professor, Head of Laboratory
Department of Neurogenetics of Language
Rockefeller University
New York, NY
Robert E. Kass, Ph.D.
Maurice Falk University Professor of Statistics and Computational Neuroscience
Department of Statistics and Data Science, Machine Learning Department and the Neuroscience Institute
Carnegie Mellon University
Pittsburgh, PA
Tuuli Lappalainen, Ph.D.
Professor, Director of the National Genomics Infrastructure and the Genomics Platform of SciLifeLab
KTH-Royal Institute of Technology
Stockholm, Sweden
Senior Associate Faculty Member
New York Genome Center
New York, NY
Cathryn M. Lewis, Ph.D.
Professor of Genetic Epidemiology & Statistics
Head of Department, Social, Genetic Developmental Psychiatry Centre
King’s College London
London, UK
Masako Suzuki, D.V.M., Ph.D.
Assistant Professor, Department of Nutrition
Texas A&M University
College Station, TX
Brenden Tervo-Clemmens, Ph.D.
Assistant Professor
Department of Psychiatry & Behavioral Sciences
Masonic Institute for the Developing Brain
University of Minnesota
Minneapolis, MN
Joshua T. Vogelstein, Ph.D.
Assistant Professor
Department of Biomedical Engineering
Johns Hopkins University
Baltimore, MD
NIMH Staff
Jonathan Pevsner, Ph.D. (co-lead)
Chief
Genomics Research Branch
Division of Neuroscience and Basic Behavioral Science
Laura Rowland, Ph.D. (co-lead)
Chief
Neuroscience of Mental Disorders and Aging Program
Division of Translational Research
Jasenka Borzan, Ph.D.
Scientific Review Officer
Division of Extramural Activities
Jeymohan Joseph, Ph.D.
Chief
Neuropathogenesis, Genetics and Therapeutics Branch
Division of AIDS Research
Susan Koester, Ph.D.
Deputy Director
Division of Neuroscience and Basic Behavioral Science
David Panchision, Ph.D.
Chief
Developmental and Genomic Neuroscience Research Branch
Division of Neuroscience and Basic Behavioral Science
Lori Scott-Sheldon, Ph.D.
Chief
Data Science and Emerging Methodologies in HIV Program
Division of AIDS Research
Tracy Waldeck, Ph.D.
Director
Division of Extramural Activities
Andrea Wijtenburg, Ph.D.
Chief
Brain Circuitry and Dynamics Program
Division of Translational Research
[Top]
II. Introduction
Recent technological advances have vastly increased the amount of data that researchers can collect. In response, the National Institute of Mental Health (NIMH) has funded studies involving much larger, more complex datasets—which, in turn, has led to new insights and important steps toward improved prevention, diagnosis, and treatment. For example, investigators can link high dimensional biological measures—such as omics and brain imaging—with behavioral, environmental, and social determinants of health measures to analyze mental health outcomes. Studies that involve epigenetics, microbiome, and genomics data are also examples of the use of high dimensional data.
The current standards for study design and analysis in research using high dimensional data are sometimes insufficient for ensuring that findings are robust, reproducible, and generalizable. For example, most links between the natural variation in biology and mental health phenotypes across populations have small effect sizes, necessitating large sample sizes and/or novel research designs. Depending on the study design and outcomes studied, effect sizes may appear large but have wide confidence intervals that limit insights into true effects. In some instances, only a handful of participants are needed to obtain compelling evidence in support of a claim (Newbold et al., 2020).
Another challenge is that access to both large datasets and substantial computational power may lead to inadvertent violations of well-established norms in data science, statistical reasoning, methodological rigor, and transparency. Thus, while the use of high dimensional data has the potential to change the way in which mental illnesses are understood, investigators must navigate specific challenges to ensure rigor and reproducibility. Investigators who use high dimensional data would therefore benefit from both general best practices and nuanced principles in their research designs. Investigators would also benefit from guidance for evaluating the robustness of science, which could be outlined in both published literature and grant applications.
[Top]
III. The Charge
To assist in prioritizing the funding of rigorous, reproducible research, NIMH convened an ad hoc workgroup, the National Advisory Mental Health Council Workgroup on High Dimensional Data (the Workgroup). The Workgroup’s charge was to develop recommendations regarding appropriate conceptual frameworks and experimental and analytic designs for studies using high dimensional datasets. NIMH proposed potential areas for the Workgroup to consider, including: 1) ensuring statistical rigor and enhancing reproducibility, 2) understanding the role of hypotheses and conceptual frameworks, 3) guiding studies involving peripheral biomarkers, and 4) assessing the clinical utility of studies. This report is organized by topic areas, which outline the challenges common to studies using high dimensional data, describe the Workgroup’s deliberations and/or rationale, and offer recommendations for both general best practices and specific considerations.
Definitions
Working definitions for key terms are used throughout this report. Such definitions are neither exhaustive nor without ambiguity, but are included to maintain consistency and clarity across the Workgroup’s recommendations. Where relevant, this report directs readers to more comprehensive discussions on both historical and current use of terms and definitions.
Workgroup Framework
The Workgroup began its deliberations with a key observation—for a study to be successful, it must precisely characterize the questions to be answered. The art of converting scientific mysteries into precisely defined, answerable questions is fundamental to all quantitative sciences. To conduct research with precision and rigor, the questions under investigation should be translated from vague statements into precise language, often using mathematics. The various types of questions being investigated may be characterized in many ways.
The Workgroup aimed to develop general best practices important for all high dimensional studies, as well as specific practices that vary by study design. To achieve this, the Workgroup first distinguished five broad categories of research design (described in detail below): 1) Exploratory or Descriptive Studies, 2) Inferential or Predictive Studies, 3) Causal Intervention Studies, 4) Mechanistic or Explanatory Studies, and 5) Methods or Tool Development. It is important to note that any given study may involve two or more of these types of designs. While studies using high dimensional data might expand to other designs not described here, the goal is that the principles outlined in this report can be applied to other domains as well.
Five Broad Research Design Categories
Exploratory or Descriptive Studies
This category comprises questions about the data, as opposed to data’s relationship to the circumstances that produced the data. These questions may require the quantification of both endogenous features (e.g., physiological, anatomical, genetic) and exogenous features (e.g., questionnaire responses, eye movements, working memory assessments), and may also involve quantification of variability across subjects. Studies that emphasize data collection often include data descriptions, but exploratory research methods generally precede inferential or predictive approaches and often lead to well-defined pre-processing steps. In some cases, convincing results may be revealed without the need for inferential or predictive statistical procedures.
Inferential or Predictive Studies
The research questions in this category involve the use of data to find associations or predictions that can apply to other circumstances. This may include statements concerning the ways that results suggest or are consistent with specific scientific interpretations or explanations. Such studies typically rely on formal procedures of statistical inference, such as confidence intervals and significance tests. Predictive studies may not always use formal statistical inference techniques. Rather, they may depend on held-out datasets and empirical claims. Nonetheless, any predictions about unseen data will either explicitly or implicitly depend on inferences, as well as the assumptions necessarily associated with those inferences. While inferential and predictive studies may have divergent goals (e.g., testing a hypothesis versus predicting an outcome), they may also rely on the same statistical foundations. Rigorous conclusions require attention to both observed and unobserved confounding variables. Therefore, there is considerable overlap in experimental design for inferential and purely predictive studies. For example, longitudinal studies, cohort studies, association studies, and risk stratification studies could be used for either inference or prediction. The distinction between the two types of studies pertains to their goals and may have distinct requirements related to power and sample size, and replication.
Causal Intervention Studies
This category involves outcomes that might occur as the result of a manipulation or intervention. The conclusions drawn from such outcomes are therefore considered causal. The justification for clinical interventions ultimately rests on studies that answer such questions and typically involve formal statistical inference procedures. Both observed and unobserved confounding variables should again be addressed. Indeed, solid causal evidence is obtained only when these confounds are appropriately addressed. Common causal intervention designs include randomized controlled trials, which are often single- or double-blinded and may include crossover designs.
Mechanistic or Explanatory Studies
The research questions in this category involve using data to understand the mechanisms by which systems, circuits, and their individual components generate phenomena of interest. The goal of mechanistic studies is to develop putative explanations that can provide explanatory insights. Scientific importance may be further enhanced when these mechanistic characterizations are coupled with successful interventional outcomes. Common designs for mechanistic studies include in vitro experiments and animal studies in which environment or genetics are manipulated, and these designs may involve formal modeling.
Methods or Tool Development
This category comprises developing new methods or tools for analyzing high dimensional data, as well as significantly improving existing methods and tools. The advancements in techniques or tools may lead to novel approaches for identifying mechanistic characterizations or assessing phenomena of interest. For example, an investigator could develop a new statistical method to predict disease outcomes from multi-omics datasets.
Terms Important for Reproducible Research
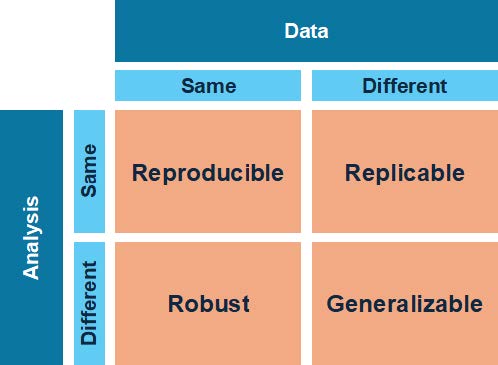
Figure 1: Depictions of reproducible, replicable, robust, and generalizable research (adapted from The Turing Way, 2022; reproduced in Leipzig et al., 2021). Reproducibility, replicability, robustness, and generalizability are key terms for defining reproducible research, and we have chosen to use them in a specific way, following Leipzig et al. (2021). Figure 1 illustrates the relationships between these key terms within a research design.
- Reproducibility: Results are reproducible when the same analysis performed on the same dataset produces the same result.
- Replicability: Results are replicable when the same analysis performed on different datasets using the same analysis workflow produces qualitatively similar answers.
- Robustness: Results are robust when different
analysis workflows (e.g., those with different statistical models, assumptions, or procedures) are applied to the same dataset, and produce similar or identical answers. - Generalizability: Results are generalizable when different analysis workflows are applied to a different dataset and produce similar or identical answers—that is, when obtaining a result does not depend on a particular dataset or analytic workflow.
Other Important Terms
There are other key terms relevant to studies using high dimensional data.
- Hypothesis: A research hypothesis proposes a tentative explanation about a phenomenon, or a narrow set of phenomena observed in the natural world. The null and alternative hypotheses provide competing answers to a research question. The null hypothesis (H0) suggests that there is no effect on the population, while the alternative hypothesis (Ha or H1) suggests that there is an effect on the population. Generally, the alternative hypothesis is the research hypothesis of interest.
- Hypothesis-driven versus Hypothesis-free Aims: Studies that aim to identify relationships or patterns using high dimensional data (within or between groups) are often described as hypothesis-driven or hypothesis-free approaches. Hypothesis-driven studies focus on a subset of features and tend to be based on prior knowledge, theories, or conceptual frameworks. Hypothesis-free or data-driven studies consider a broad range of all of the features that can identify unforeseen relationships in the data.
- Statistical Power or “Power”: The likelihood of a hypothesis test detecting a true effect (via statistical significance testing at a given threshold) if one exists. It is the probability that a test will correctly reject a false null hypothesis. Power is calculated based on sample size (n), effect size (d), significance level (á; the threshold for rejecting the null hypothesis), and the null and alternative distributions.
- Effect Size: Effect size is a quantitative measure of the magnitude of an experimental effect. It can refer to the raw difference between group means (i.e., absolute effect size) or standardized measures of effect, such as Cohen’s d or z-score.
- Test, Training, and Validation: These terms are commonly used in machine-learning studies. A training dataset is used to train or fit a machine-learning model. A validation dataset is used to evaluate the model performance during the process of tuning model hyperparameters. A test dataset is a set of data used to provide an unbiased evaluation of the final model fit on the training dataset (adapted from Shah, 2020; Acharya, 2023; and Brownlee, 2020).
[Top]
IV. General Recommendations by Topic Area
Topic 1: Goals, Aims, and Hypotheses
In science, hypotheses can be useful—sometimes essential—for formulating conceptions such that they allow specific outcomes to provide evidence about them. This can help clarify the goals and aims of a study. In some cases, hypotheses can still be useful even when they are not precisely stated. However, when precisely stated, they can determine the number and complexity of statistical test procedures needed in an analysis.
In analyses of high dimensional data, hypotheses can also narrow the search space among high dimensional features (e.g., genes or brain regions). This can influence research decisions related to setting thresholds for statistical significance, such as corrections for multiple comparisons.
The Workgroup acknowledged that it might be appropriate and valuable for transparency to use more lenient statistical significance thresholds for hypothesis-driven analyses compared to more exploratory “hypothesis-free” analyses. However, the validity of such applications can be challenging to verify. Some Workgroup members suggested implementing universal requirements for multiple comparison-adjusted significance levels—particularly in studies using whole genome or whole brain data—to reflect the fully acquired high dimensional dataset, even when a study claims to examine a specific hypothesis. Other Workgroup members felt such recommendations might overlook important context and could shift focus from the investigator’s essential goal of demonstrating to reviewers that they have addressed fundamental issues in data analysis, particularly in terms of procedures to improve the replicability of findings.
- Recommendation: All aspects of research designs, including sample size, should be well justified. See also Topic 3: Power and Sample Size.
- Recommendation: Pre-registration of hypotheses is encouraged—see Topic 9: Publication Plan. For studies involving human clinical trials, this can be done at clinicaltrials.gov.
- Recommendation: Results concerning hypotheses that are determined from exploratory analyses should be replicated. See Topic 6: Independent Replication, and Generalizability of Results.
- Recommendation: The primary goal of high dimensional predictive studies should be effective prediction. However, effects of specific variables may be more difficult to establish. In the absence of replication in separate datasets, identifying such effects may be impossible or require specialized techniques.
Topic 2: Experimental Design
Mental health research involving high dimensional data employs a broad range of experimental design, including studies of human brain activity and behavior, post-mortem tissues, in vitro cell-based and animal models, as well as studies of multi-omic genetic, transcriptomic, epigenomic, and phenotypic data.
- Recommendation: Investigators should clearly state their proposed experimental design and their justification for it in their grant application. It is important that the study design (i.e., variables, outcomes, methods, experiments) either tests specific hypotheses or provides a justification for a hypothesis-free approach, such as for exploratory or descriptive studies.
- Recommendation: When data are collected with the expectation of using inferential, predictive, or interventional procedures, the study design should be aligned with this objective. For instance, when making an inference, the sample size should be large enough to provide solid evidence of a meaningful effect, if one exists. This requires an understanding of what constitutes a meaningful effect size, whether in clinical practice or within a scientific theory. Similarly, for prediction, the sample must be large enough to exceed a meaningful criterion of predictive accuracy, if the predictor (such as a biomarker) is truly effective. This also requires knowledge of what constitutes meaningful accuracy (see Topic 3: Power and Sample Size Estimates).
- Recommendation: Investigators should clearly indicate 1) the goal of the experiment (i.e., which questions the data and analyses will answer), and 2) justification of the validity of their experimental design, including both data acquisition and analysis. A design is valid with respect to the goal if there is a high likelihood that the design will yield data and analyses that address the goal. Preliminary or simulated results can provide compelling evidence that the design will address the original stated goal.
Topic 3: Power and Sample Size Estimates
Statistical power is essential to hypothesis testing and therefore relevant for inferential-predictive, causal-interventional, and mechanistic studies. While statistical power may not be relevant to purely descriptive studies, statistical methods are routinely applied to such studies, nonetheless. Statistical power is the probability that a test will correctly reject a null hypothesis when it is actually false. Power is calculated based on sample size (n), effect size (d), standard deviation of the outcome(s), significance level (á; the threshold for rejecting the null hypothesis).
The Workgroup identified several notable challenges with the current use of power analyses in applications for high dimensional data analyses, including: 1) relevant effect sizes and estimates of standard deviation used in power analyses should be derived, ideally, from multiple well-powered prior benchmarking studies and 2) closed-form/analytic power calculations are often not available for more complex high dimensional multivariate analytic techniques (e.g., “machine learning”), necessitating more complex simulation-based approaches. In some cases, power analyses may not be feasible (e.g., because of the assumptions that are required when data from previous, similar studies are not available).
The Workgroup determined that large sample sizes are generally necessary in population-level association studies (e.g., brain-wide association studies), which are often exploratory-descriptive, and in studies for which small effects sizes are typical. In contrast, more descriptive studies of central tendencies, group-average descriptions, and some interventional studies often do not require large samples—assuming there is use of reliable and valid measurements. For example, Gratton et al. (2022) discusses meaningful discoveries being made with large or small sample sizes.
The Workgroup noted that genome-wide association studies sometimes aggregate multiple studies that would otherwise be underpowered on their own. The Workgroup emphasized the importance of generating resources that in isolation may be unpowered, but that could be combined with others—particularly for new but rapidly expanding datatypes that contribute to a larger set of analysis. More emphasis should be placed on explaining the choice of the sample set and, in case/control studies, how cases and controls are matched.
Additionally, the Workgroup discussed the importance of explicit training, guidance, and consideration for trainees and early career researchers, whose projects and resources are often smaller in scale. Leveraging existing well-powered datasets for inferential and prediction studies, and designs examining central tendencies, or interventions that do not require large samples for training awards that require data collection, was discussed as a strategy in the current funding environment (see Topic 10: Early Stage Investigators and Training Grants).
- Recommendation: The power calculation in a grant application should be based on known, empirically-generated effect sizes and standard errors or percent variance explained. Examples of well-powered studies may be found in conjunction with NIH supported open repositories, the NIH Brain Research Through Advancing Innovative Neurotechnologies® (BRAIN) Initiative, hiPSC technology1, and the Adolescent Brain Cognitive Development Study (ABCD Study®).
- Recommendation: Where previous sufficiently powered data exist, the choice of effect size should be well documented in the grant application by referencing specific figures or tables found in published literature. Investigators should also acknowledge whether the strongest effect observed is the more commonly observed effect.
- Recommendation: When there are no previous sufficiently powered data available for investigators to reference, power analyses should consider a range of plausible estimates of effect sizes, measurement reliability, and attrition/available samples. Related well-powered studies can be used to guide the consideration of ranges.
- Recommendation: If the data type is novel and there is no prior information available, then it may help reviewers to define the percentage variance explained or effect size detectable with the proposed sample size through analogies to other well-powered data or effects from other fields. In this case, estimates should be conservative and not based on a best-case scenario. For example:
- If an investigator plans to conduct a novel cross-sectional study of functional brain associations with a new behavioral phenotypic measure for which no data exist in the literature, the Workgroup recommends that the study be sufficiently powered to detect effects similar to those observed in other behavioral measures from large well-powered samples (e.g., univariate r~0-0.2 in ABCD Study, UK Biobank). Unless otherwise proven, power and sample size estimates should be based on this range from prior related data, rather than assuming larger or even top values within this range.
- Recommendation: Investigators should consider using well-justified and appropriately documented simulation approaches to aid in power analyses that are tailored to study specific questions and novel high dimensional data analyses. Simulations would be strongest when including a range of potential study outcomes, including effect size, sample size, and measurement reliability. These procedures can protect against an optimistic bias or a “just powered study” that lacks robustness to even minor deviations in effect size, sample size, and reliability.
- Recommendation: Power analyses should appropriately match the proposed research design and statistical methodology. For study designs that examine within-subject effects or include an intervention or manipulation (e.g., interventional/mechanistic), fewer samples may be needed compared to traditional exploratory or descriptive designs. This is due to reduced measurement error of between-subject effects and the estimation of potentially larger within-subject or longitudinal effects. Similarly, predictive analyses focused solely on predicting an outcome—rather than the reliability and stability of the individual features contributing to the prediction—will likely require fewer and more distinct samples than exploratory or observational designs. However, it is important to note that not all Workgroup members shared this view. Study designs with relatively smaller sample sizes should clearly justify how the smaller number of samples is sufficient and appropriately powered to answer the research question, such as by capturing larger effect size longitudinally.
- Recommendation: When reporting power calculations, investigators should provide information about the known variance in their measurements, including test-retest measurement reliability, when available.
Topic 4: Measurements, Traits, and Phenotypes
The Workgroup’s charge included a wide range of high dimensional biological data from various biological source materials, including brain imaging, genomics, peripheral biomarkers, epigenetics, and the microbiome. The Workgroup emphasized the importance of having reliable, validated, and interpretable measurement for each type of data. Investigators are encouraged to stay informed about emerging evidence and field standards for each data type.
The quality of psychiatric trait/phenotype data is a key determinant of the rigor and reliability of analyses that seek to link them with high dimensional data. The Workgroup noted that, historically, psychiatric phenotype reliability and validity may have been de-emphasized in applications focused on high dimensional biological data.
Just as larger sample sizes can increase statistical power, increasing measurement precision can also increase statistical power. For example, Nebe et al. (2023) described determinants of precision for a range of neuroscience applications (magnetic resonance imaging [MRI], magnetoencephalography [MEG], electroencephalography [EEG], eye-tracking, endocrinology) and provided guidance on increasing reproducibility. However, while technical variation can be reduced, it is rarely entirely removed. This can lead to false positive discoveries if not accounted for, particularly if technical batch effects are confounded with biological variables of interest.
- Recommendation: Studies should include sufficient biological and technical replicates, where applicable, and investigators should describe how the robustness of data measurements will be evaluated.
- Recommendation: Investigators should establish how technical factors involved in obtaining biospecimens and generating data can introduce variation and affect the interpretability of outcomes. These factors include tissue collection, biomolecule extraction, and assay execution. Although using consistent protocols reduces technical variation, technical batch effects are often unavoidable. Studies should consider and describe strategies to mitigate their potential confounding impact.
- Recommendation: Unless otherwise justified, existing standards on data capture, cleaning, processing, and analytics should be followed. Investigators should consider potentially relevant biases that can be introduced even in standardized raw data processing steps (e.g., sequencing read alignment).
- Recommendation: Investigators should discuss the relative strengths and limitations of psychiatric phenotype measurements, even for commonly used measures.
- Recommendation: When appropriately evaluated, new approaches to phenotyping can improve reproducibility and rigor. These approaches may include multi-assessment and multi-informant approaches, as well as novel data sources such as electronic medical records, wearables, and high-throughput survey designs (e.g., ecological momentary assessment).
- Recommendation: Due to practical limitations, many studies must use biospecimens that are imperfect proxies of the actual tissues and cell types of interest. Investigators should describe how processes measured in the proposed biospecimens will relate to disorders and traits of interest, as well as how their findings will be validated. When measuring bulk tissue samples, investigators should consider how variability in cell type composition can affect the results and how it will be addressed.
Topic 5: Dimensionality Reduction
The Workgroup acknowledged that high dimensional data are subject to a “curse of dimensionality” and many methods of variable selection and other dimensionality reduction strategies are routinely applied. Although having a large number of variables can provide new and interesting opportunities, contemporary analytical methods often rely on sophisticated algorithms to select among variables or to combine their effects. Established algorithms provide well-defined procedures and many have known theoretical properties. While dimensionality reduction is not always necessary, if investigators choose to reduce the dimensionality of the data using data-driven approaches (as opposed to hypothesis-based or biologically-based approaches), it is important to follow approaches that ensure rigor and reproducibility.
The Workgroup also discussed analytic methods that produce a “best” set of variables or a “best” way to combine them. They noted that there is little evidence that a putatively “best set” is substantially better than dozens or hundreds of alternative choices, which largely depends on how the model is trained and tested and the criterion used to define “best”. Thus, interpretation of an analysis may heavily depend on the chosen “best set” of variables and may not represent appropriate conclusions to be drawn from the data. Substantive conclusions will be convincing for the high dimensional data only if they remain consistent across the many plausible statistical modeling candidates. Otherwise, the conclusions are only reflective of the specific analysis and statistical inference conducted and should be interpreted clearly as such. Developing advanced statistical methods for dimensionality reduction is an active and fruitful area of research.
- Recommendation: To enhance rigor of dimensionality reduction methods as applied to high dimensional research, investigators should justify why a particular method is applied to the data and how it will lead to valuable conclusions.
- Recommendation: Investigators should provide specific details of the dimensionality reduction method to ensure that the same approach could be applied independently to external testing or validation data by other investigators.
- Recommendation: Investigators should select a variable set, or a way to combine the variables, to optimize a specific objective. Investigators should qualify their interpretations based on a data-driven reduced dimensionality variable set by referencing this specific objective.
Topic 6: Independent Replication, and Generalizability of Results
The Workgroup emphasized the importance of evaluating the reproducibility, robustness, replicability, and/or generalizability of key results to analytical choices such as data processing, parameter selection, and alternative statistical methods. This is particularly important for methods and data types where gold-standard analysis methods are not yet well-established. The use of terms related to reproducibility, robustness, replication, and generalizability may vary by field and study design, and their implementation and analyses may differ accordingly. See Definitions (above) for more information.
The Workgroup also highlighted the value of existing large-scale biobanks, including those funded by NIMH, as critical resources to test robustness, replicability, and generalizability. In some cases, robustness, replicability, and generalizability may not be immediately achievable but should remain a future goal. Therefore, it is essential to carefully consider statistical procedures, such as those used in statistical inference.
While there is utility in adhering to established reporting standards, such as conventional levels of significance, these standards serve only as rough guides for interpretation. The scientific importance of statistical results must be evaluated in relation to the broad body of knowledge that defines their context. For example, in a gene expression study, a transcript may be statistically significantly regulated with a small fold-change that implies lack of biological significance in isolation. Only in a broader context might such a finding and small effect reveal clinical significance.
Statistical findings must also consider the totality of study procedures. Analysts often examine complex data in various ways, using different pre-processing steps, as well as exploratory plots and summaries, for quality control purposes and in order to better understand what the data might have to offer. While this is encouraged, formal statistical procedures, such as confidence intervals and hypothesis tests, typically do not account for these preliminary efforts. In some cases, it can be argued that certain pre-processing steps are both standard and unlikely to alter the properties of statistical procedures. However, extensive data explorations without pre-planned or “pre-registered” procedures can undermine generalizability and ultimately the interpretations of the outcomes. Similarly, when predictive methods include preliminary steps that are tuned to the predicted data (i.e., overfitting), accuracy will appear inflated, leading to erroneous conclusions.
Some investigators undertake extensive exploration of data processing and analysis, and selectively focus on those statistical tests that support their hypothesis. Such an approach undermines assertions about statistical results. This is known as p-hacking. P-hacking can undermine the standard interpretation of p-values. Stefan and Schonbrodt (2023) enumerated many forms of p-hacking. It is important to note that the use of confidence intervals or Bayesian methods does not automatically prevent invalid interpretations resulting from multiple passes through the data.
The risk of corrupted inferences due to preliminary analysis is one reason many statisticians often hesitate to take results as solid evidence—unless those results have very small p-values, which are more likely to represent replicable deviations from null hypotheses. A good way to guard against this type of inference distortion is to replicate with fresh data after “freezing” and pre-specifying all data-analytic procedures. However, this does not eliminate the possibility of bias (see, for example, Rosenbaum, 2001). In particular, causal claims (which concern interventional questions) almost always require satisfying conditions for statistical causal inference, based on randomized controlled trials or other methods.
- Recommendation: Best practices include plans for replication and tests of generalizability in projects relying on high dimensional data. For example, predictive studies should validate developed algorithms using independent data, without developing any aspects of the model, in order to prevent algorithmic overfitting.
- Recommendation: In meta-analysis, for which the goal is to combine all possible studies to achieve the highest power, investigators are encouraged to test for comparability of results across included studies. Follow-up experimental work to test mechanistic hypotheses provides further important support.
- Recommendation: Investigators studying general associations between high dimensional biological data and psychiatric traits should emphasize how they address generalizability—for example, to determine whether the observed associations in a given sociodemographic group can be generalized to other sociodemographic groups. Note that if the question is about generalizing to other groups, it is not exploratory anymore—it is inferential. In contrast, interventional studies may have limited ability to test results in a new sample due to complex and/or novel designs. However, single-participant or n=1 (“idiographic”) analyses that replicate patterns and/or inferential statistics across individual participants may also be useful.
- Recommendation: In high dimensional data analyses, investigators should prioritize, when possible, efforts to test the replication of results to either an independent set of data from the same samples or to a new fully independent sample set.
- Recommendation: Investigators should include explicit plans to test for robustness, replication, and generalizability.
- Recommendation: Promoting robustness, replication, and generalizability can be supported by pre-registering hypotheses and sharing data, metadata (Leipzig et al., 2021), and computational workflows. Additionally, adopting other best practices described in Kass et al. (2016) and Chen et al. (2019) are encouraged in application plans.
- Recommendation: Investigators should share bioinformatic pipelines used to analyze high dimensional data as these face particular challenges in ensuring reproducible research. For more information, see Topic 8: Data, Code, and Resource Sharing.
Topic 7: Pilot Studies/Preliminary Data
Due to the nature of high dimensional data and expected complexity in identifying biomarkers, pilot studies with small samples are unlikely to be adequately powered. Pilot studies on high dimensional data will be most useful towards demonstrating feasibility, technical reproducibility, and safety (if relevant) of the proposed data collection procedures and analyses. The Working Group acknowledged that, despite these limitations, pilot studies are an important prerequisite of larger and well-powered studies, and thus a valuable component of high-dimensional data.
- Recommendation: In pilot studies and preliminary data, measures of effect sizes and statistical significance testing should be approached with a considerable degree of caution. Power analyses for high dimensional data based on small pilot studies could be avoided in favor of a range of plausible estimates based on the well-powered recent work and emerging field standards.
Topic 8: Data, Code, and Resource Sharing
The Workgroup emphasized the value of existing standards for open science supported by NIMH, as well as the particular importance of data, metadata, code, and software sharing. Additionally, investigators should follow both the principles of Findability, Accessibility, Interoperability, and Reusability (FAIR) (Wilkonson et al., 2016; Barker et al., 2022) and Transparency, Responsibility, User Focus, Sustainability and Technology (TRUST) (Lin et al., 2020). This fosters reproducible computational research, enables reuse of the valuable data for further biological questions and methods development, and enables later studies that integrate data from multiple sources for better statistical power, diverse study populations, and/or multimodal insights. The scientific value of data reuse is only going to grow with analytical methods development.
Metadata provide crucial information about data, including its source, format, context, content, and control. Metadata describe the data, offer quality control information, are necessary for integration with other datasets, guide the use of data, and facilitate data sharing. Leipzig et al. (2021) discussed the importance of metadata standards. Ziemann et al. (2023) recommended a framework for reproducible computational research involving high dimensional data. This framework includes five components: 1) literate programming, 2) code version control and persistent sharing, 3) compute environment control, 4) persistent data sharing, and 5) documentation.
- Recommendation: Investigators should develop a detailed data, code, and resource sharing plan that includes: 1) the types and amounts of data to be generated; 2) the data repositories where data will be found; 3) the biological repositories where biospecimens (e.g., DNA or cell lines) will be made available; 4) and a detailed plan for code sharing. Specifically:
- Investigators should comply with NIH data sharing policy and NIMH data sharing policy (NOT-MH-23-100 ) to make raw and processed data available to the research community.
- NIMH requires human subjects data to be shared with the NIMH Data Archive (NDA) for specific awards.
- A detailed data access plan should be provided for studies using retrospective data.
- Investigators should comply with NIMH requirements to ensure that all biospecimens are consented appropriately to facilitate wide-scale distribution.
- Plasmids and libraries should be made available through established resources such as Addgene .
- All new and engineered cell lines should be deposited in a repository such as the NIMH Repository and Genomics Resource (NRGR) . Conditions for expansion and/or differentiation of cell lines should be clearly documented, and cell line authentication should be transparent and include genotyping markers.
- NIMH requires that human DNA and stem cell lines be deposited within the
NRGR , in compliance with the biospecimen sharing policy . - Investigators should include a plan for sharing metadata, code, and analysis.
- A detailed plan for code sharing should include standards and code documentation (e.g., Kiar et al., 2023). Other investigators should be able to run the data codes and obtain the same published results without having to contact the author.
- When possible, open repositories such as GitHub , GitLab and BitBucket should be used.
- Recommendation: In adhering to NIH data sharing policy , investigators should provide up-to-date data, code, and resource sharing plans and/or their availability in all progress reports, pre-prints, and publications.
- Data, code, and resources should be easily searchable using a journal’s Digital Object Identifier (DOI).
- Data should be stored in appropriate repositories to facilitate reproducible research, along with information about their location and accessibility.
Topic 9: Publication Plan
Many investigators alter their research design and analysis plans in the middle of their studies, and this may be especially true for studies involving high dimensional and big data. There are many reasons to make alterations, such as accounting for preliminary observations or adjusting sample size during an experiment. However, there are also concerns associated with altering research design and analysis plans. For instance, these changes can lead to p-hacking, in which more favorable outcomes are achieved through selective choice of statistical tests or selective reporting of significant results. Another concern is revising hypotheses to match results, a practice known as HARKing ("hypothesizing after the results are known”).
A publication plan outlines efforts to maximize transparency and reduce these concerning practices. Part of a publication plan may include efforts to pre-register hypotheses, analytic methods, and/or data-driven analyses and goals. While no specific platforms for pre-registration are endorsed by the Workgroup, some examples include Open Science Framework, Center for Open Science, and Zenodo.
Pre-registration involves creating a time-stamped document before collecting and/or analyzing data (possibly after an embargo period) that can be published in a journal or shared in a public repository. Versioning allows for any adjustments or changes that may occur to be documented and compared with the final manuscript. Pre-registration has been shown to increase the transparency of research and reduce questionable research practices. However, the practice has been less common in some research areas focused on high dimensional data.
Other components of the publication plan might include efforts to mitigate publication bias. Publication bias occurs when certain research findings, such as positive results, are more likely to be published than other findings, such as negative results. This can distort the knowledge base in a field because the full range of research knowledge is not represented in the published literature. Publication bias can also adversely impact study design by encouraging investigators to focus on specific positive results or engage in p-value hacking. One negative consequence of publication bias is that meta-analyses may overestimate true effect sizes because they are biased toward analyzing results reported as positive. Conclusions drawn from such meta-analyses may be skewed, with biased samples of data leading to data summaries characterized by excess heterogeneity due to larger effect sizes.
Practices such as including null findings in supplemental materials of a published manuscript or in a preprint can help disseminate a scope of research results. Preprints are early versions of an unpublished manuscript released on a preprint server such as bioRxiv, arXiv, and OSF Preprints, which can be later updated with newer versions. Publishing on preprint servers increases the discoverability of results; alters the incentive structure of academic publishing to favor novel findings; and may counteract some undesired effects of big data, high dimensional data, and open access datasets (e.g., opportunities for a multitude of hypotheses and/or statistical and computational approaches that may increase unreproducible research). Preprints also provide a relatively fast and easy way to share preliminary work, which can facilitate feedback from peers and enhance transparency of the iterative nature of research (Verma et al., 2020).
Although pre-prints are not peer reviewed, many journals encourage preprints and will accept manuscripts that have been previously posted on a preprint server. Preprints on eligible servers that acknowledge NIH funding are also indexed by PubMed Central and PubMed .
- Recommendation: Pre-registration is unique to each research project. Although pre-registration is not required, inclusion of a pre-registration plan is considered a strength of the research approach and publication plan.
- Recommendation: Plans to publish all findings, not just a subset of the results, as well as null results are strongly encouraged. Papers on high-dimensional data should typically include figures and statistics describing the full distribution of p-values and other test statistics, even if individual discoveries are characterized further via follow-up analyses and experiments.
- Recommendation: Preprints are a good solution to publish comprehensive findings, especially null findings.
- Recommendation: All publications should comply with NIH public access policy.
- Recommendation: Publication in predatory journals should be avoided (See: https://beallslist.net ).
Topic 10: Early Stage Investigators and Training Grants
Many early-stage researchers, such as postdoctoral fellows, training awardees, and early-stage faculty, have limited financial resources to design projects that generate large, well-powered, high dimensional datasets. This is because non-R01 NIH funding mechanisms for relatively early-stage scientists typically have lower maximum per year funding levels and may be of shorter duration, making it impractical to generate expensive datasets. Ideally, early-stage investigators would have the ability to 1) generate their own data, if part of their goals and 2) pose and answer questions from data (either their own or generated by others).
- Recommendation: Leveraging published data from individual researchers or repositories for smaller awards is highly encouraged.
- Recommendation: There should also be support for training investigators to collect their own data. Investigators could combine newly generated data with existing large datasets to increase sample size. Alternatively, investigators could seek alternative study designs that are informative and sufficiently powered with smaller scale data, although these types of data may be newer and viewed as riskier. Investigators should be given resources such as NIH Toolbox to harmonize data acquisition with larger, existing datasets.
- Recommendation: In some cases, such as training awards, the budget may be too limited to allow inclusion of a large sample size, and therefore the study may be underpowered. Reviewers should take into consideration the training purpose of the proposed work, and some cases choose to support the funding of underpowered studies.
[Top]
V. Conclusion
This report provides an overview of many challenges that investigators face when conducting research using high dimensional data. The Workgroup emphasizes the need for robust, reproducible research—outlining best practices and specific considerations for study design, power and sample size estimates, measurements of traits and phenotypes, dimensionality reduction, replication, and generalizability. In their report, they also address the importance of data, code, and resource sharing to enhance transparency and reproducibility. Additionally, the Workgroup discussed the role of pilot studies, publication plans, and support for early-stage investigators.
The Workgroup’s recommendations aim to guide both investigators and NIMH toward rigorous and reproducible research outcomes in the context of high dimensional data analysis. Many components of this guide are applicable to research generally. By adopting these best practices, investigators can leverage high dimensional data to uncover new insights and improve the diagnosis, treatment, and prevention of mental illnesses.
[Top]
VI. References
Acharya, A. (2023, June 13). Training, Validation, Test Split for Machine Learning Datasets. Encord.com. https://encord.com/blog/train-val-test-split/
Barker, M., Chue Hong, N. P., Katz, D. S., Lamprecht, A.-L., Martinez-Ortiz, C., Psomopoulos, F., Harrow, J., Castro, L. J., Gruenpeter, M., Martinez, P. A., & Honeyman, T. (2022). Introducing the FAIR Principles for research software. Scientific Data, 9(1). https://doi.org/10.1038/s41597-022-01710-x
Brownlee, J. (2017, July 26). What is the Difference Between Test and Validation Datasets? Machine Learning Mastery. https://machinelearningmastery.com/difference-test-validation-datasets/
Chen, X., Dallmeier-Tiessen, S., Dasler, R., Feger, S., Fokianos, P., Gonzalez, J. B., Hirvonsalo, H., Kousidis, D., Lavasa, A., Mele, S., Rodriguez, D. R., Šimko, T., Smith, T., Trisovic, A., Trzcinska, A., Tsanaktsidis, I., Zimmermann, M., Cranmer, K., Heinrich, L., & Watts, G. (2018). Open is not enough. Nature Physics, 15(2), 113–119. https://doi.org/10.1038/s41567-018-0342-2
Gratton, C., Nelson, S. M., & Gordon, E. M. (2022). Brain-behavior correlations: Two paths toward reliability. Neuron, 110(9), 1446–1449. https://doi.org/10.1016/j.neuron.2022.04.018
Kass, R. E., Caffo, B. S., Davidian, M., Meng, X.-L., Yu, B., & Reid, N. (2016). Ten Simple Rules for Effective Statistical Practice. PLOS Computational Biology, 12(6), e1004961. https://doi.org/10.1371/journal.pcbi.1004961
Kiar, G., Clucas, J., Feczko, E., Goncalves, M., Dorota Jarecka, Markiewicz, C. J., Halchenko, Y. O., Hermosillo, R., Li, X., Miranda-Dominguez, O., Ghosh, S., Poldrack, R. A., Satterthwaite, T. D., Milham, M. P., & Fair, D. (2023). Align with the NMIND consortium for better neuroimaging. Nature Human Behaviour, 7(7), 1027–1028. https://doi.org/10.1038/s41562-023-01647-0
Leipzig, J., Nüst, D., Hoyt, C. T., Ram, K., & Greenberg, J. (2021). The role of metadata in reproducible computational research. Patterns, 2(9), 100322. https://doi.org/10.1016/j.patter.2021.100322
Lin, D., Crabtree, J., Dillo, I., Downs, R. R., Edmunds, R., Giaretta, D., De Giusti, M., L’Hours, H., Hugo, W., Jenkyns, R., Khodiyar, V., Martone, M. E., Mokrane, M., Navale, V., Petters, J., Sierman, B., Sokolova, D. V., Stockhause, M., & Westbrook, J. (2020). The TRUST Principles for digital repositories. Scientific Data, 7(1). https://doi.org/10.1038/s41597-020-0486-7
Nebe, S., Reutter, M., Baker, D. H., Bölte, J., Domes, G., Gamer, M., Gärtner, A., Gießing, C., Gurr, C., Hilger, K., Jawinski, P., Kulke, L., Lischke, A., Markett, S., Meier, M., Merz, C. J., Popov, T., Puhlmann, L. M., Quintana, D. S., & Schäfer, T. (2023). Enhancing precision in human neuroscience. ELife, 12, e85980. https://doi.org/10.7554/eLife.85980
Newbold, D. J., Laumann, T. O., Hoyt, C. R., Hampton, J. M., Montez, D. F., Raut, R. V., Ortega, M., Mitra, A., Nielsen, A. N., Miller, D. B., Adeyemo, B., Nguyen, A. L., Scheidter, K. M., Tanenbaum, A. B., Van, A. N., Marek, S., Schlaggar, B. L., Carter, A. R., Greene, D. J., & Gordon, E. M. (2020). Plasticity and Spontaneous Activity Pulses in Disused Human Brain Circuits. Neuron, 107(3), 580-589.e6. https://doi.org/10.1016/j.neuron.2020.05.007
Rosenbaum, P. R. (2001). Replicating Effects and Biases. The American Statistician, 55(3), 223–227. https://doi.org/10.1198/000313001317098220
Shah, T. (2017, December 6). About Train, Validation and Test Sets in Machine Learning. Towards Data Science; Towards Data Science. https://towardsdatascience.com/train-validation-and-test-sets-72cb40cba9e7
Spitzer, L., & Mueller, S. (2023). Registered report: Survey on attitudes and experiences regarding preregistration in psychological research. PLOS ONE, 18(3), e0281086. https://doi.org/10.1371/journal.pone.0281086
Stefan, A. M., & Schönbrodt, F. D. (2023). Big little lies: a compendium and simulation of p-hacking strategies. Royal Society Open Science, 10(2), 220346. https://doi.org/10.1098/rsos.220346
Turing Way Community, Arnold, B., Bowler, L., Gibson, S., Herterich, P., Higman, R., Krystalli, A., Morley, A., O’Reilly, M., & Whitaker, K. (2019, March 25). The Turing Way: A Handbook for Reproducible Data Science. Zenodo. https://zenodo.org/records/3233986
Verma, A. A., & Detsky, A. S. (2020). Preprints: a Timely Counterbalance for Big Data–Driven Research. Journal of General Internal Medicine, 35(7), 2179–2181. https://doi.org/10.1007/s11606-020-05746-w
Wilkinson, M. D., Dumontier, M., Aalbersberg, Ij. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J.-W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., & Gonzalez-Beltran, A. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3(1). https://doi.org/10.1038/sdata.2016.18
Ziemann, M., Poulain, P., & Bora, A. (2023). The five pillars of computational reproducibility: bioinformatics and beyond. Briefings in Bioinformatics, 24(6). https://doi.org/10.1093/bib/bbad375
[Top]
